attach23
attach23.docx
National Epidemiologic Survey on Alcohol and Related Conditions-III (NIAAA)
attach23
OMB: 0925-0628
ATTACHMENT 23
SAMPLE SIZE REQUIREMENTS FOR GENETIC ANALYSES
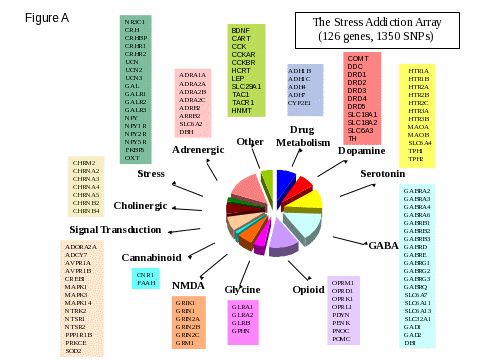
Numerous genetic variants (primarily SNP or related variants) previously identified as being related to alcohol use disorders and their associated disabilities appear in Figure A. These gene frequencies will form the basis of sample size calculations we can expect for any newly discovered or other genetic variants examined in the NESARC-III (such as CNVs). As can be seen in Table 1, these markers vary greatly in terms of their frequency, and the same marker may differ substantially in frequency within populations of different ancestry (Table 2). Second, sample size requirements for determining gene-environment or gene-gene interactions are substantially greater than those for detecting environmental or genetic effects alone. Finally, many phenotypes (outcomes) of interest have low lifetime prevalences, and a large sample size is required to ascertain a main genetic effect of large magnitude, i.e., a large odds ratio (OR). Typically, gene-disease associations between these variants have been empirically shown in the range of OR = 1.2 to 2.0. However, the current and major focus of gene-disease association studies is to identify CNVs and related variants (beyond SNPs) that relate to alcohol use disorders and their associated disabilities, and these variants are anticipated to be much more penetrant than SNPs, that is, have greater associations (OR >5.0) with the disease outcomes of the NESARC-III. To be conservative, we have retained the lower and more modest levels of associations previously found for SNP and related variants (OR = 1.2 to 2.0) in our sample size calculations. Accordingly, the required sample sizes shown in our calculations are very likely to be overestimates.
As illustrated in Figure B, for a condition such as alcohol dependence, depression or drug use disorders, all of which have a lifetime prevalence of approximately 13%, the sample size required to determine an OR of 1.2 (p<.05) in association with a given genetic variant (absent gene-environment or gene-gene interactions) varies from more than 40,000 for genetic variants with frequencies of only 5% to approximately 8,000 for genetic variants with frequencies of 50%. Associations of greater magnitude, e.g. an OR of 1.5, can be established with far fewer cases, less than 10,000 for even rare genetic variants. However, the sample size requirements to ascertain an OR of 1.5 for a genetic variant for alcohol dependence are far greater when the intent is to examine gene-environment interactions, as illustrated in Figure C. For a relatively rare environmental factor with a prevalence of only 10%, a sample size of approximately 18,000 is required if the OR for the gene-environment interaction is assumed to be 2.0. For more common environmental factors whose prevalence is 25% or greater, the sample size requirement for an interaction OR of 2.0 drops to less than 10,000. The sample size calculations in Figure C also pertain to gene-gene interactions. The sample size of the NESARC III is thus sufficient for all of these purposes.
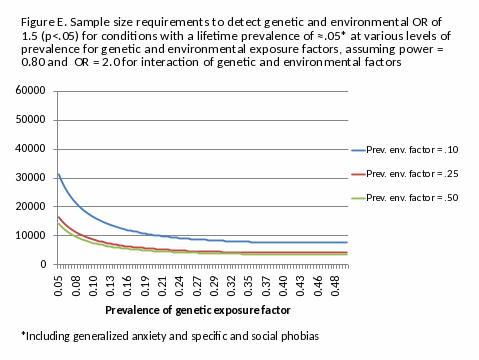
Sample size requirements are greater for outcomes with lower lifetime prevalence rates, such as most mood, anxiety and personality conditions. Figure D illustrates sample size requirements for conditions with a lifetime prevalence of 5%, which include, for example, generalized anxiety and specific and social phobias. The NESARC III sample will be adequate to determine fairly robust genetic associations (OR ≥ 1.5) for even rare genetic markers, and to determine more modest associations (e.g., OR = 1.2) with genetic markers whose prevalence is in excess of 12%, in the absence of gene-environment interactions. As illustrated in Figure E, for gene-environment interactions with OR=2.0, it will also be able to determine a genetic OR of 1.5 for almost all gene frequencies where the prevalence of the environmental factor is ≥10%.
Importantly, it should be noted again that CNVs and other unexplored gene variants are expected to be more penetrant and thus more strongly related to alcohol use disorders and their associated disabilities than are most of the currently identified genetic variants. Thus, the higher OR expected from these new gene variants will result in much smaller sample size requirements than those illustrated in Figures B to E.
Table 1. Selected allele frequencies for genetic markers of various conditions and behaviors |
||||
Gene |
Marker |
Allele |
Frequency/ Prevalence* |
Associated condition/behavior |
ADH1B |
rs1229984 |
[A/G] |
0.322 |
Alcohol metabolism |
ADH1C |
rs698 |
[A/G] |
0.39 |
Alcohol metabolism |
ADH4 |
rs1042364 |
[A/G] |
0.098 |
Alcohol metabolism |
ADH7 |
rs1573496 |
[C/G] |
0.026 |
Alcohol metabolism |
ALDH2 |
rs671 |
[A/G] |
0.082 |
Alcohol metabolism |
NPY |
rs16139 |
[A/C] |
0.067 |
Stress regulation |
SLC6A4 |
rs1042173 |
[A/G] |
0.478 |
Stress regulation, obsessive-compulsive disorder |
COMT |
rs4680 |
[A/G] |
0.337 |
Stress regulation and behavioral dyscontrol, impulsivity, attention-deficit/ hyperactivity disorder, mood disorders |
BDNF |
rs6265 |
[A/G] |
0.251 |
Stress regulation, attention-deficit /hyperactivity disorder, mood disorders |
HTR2C |
rs6318 |
[C/G] |
0.199 |
Stress regulation, behavioral dyscontrol, borderline PD, suicidality, anorexia |
OPRM1 |
rs1799971 |
[A/G] |
0.167 |
Stress regulation |
HTTLPR |
Vntr |
LG |
0.09-0.15 |
Behavioral dyscontrol, impulsivity, suicidality, mood disorders |
MAOA |
Vntr |
3.5 and 5 repeats |
0.006-0.016 |
Behavioral dyscontrol, impulsivity, antisocial personality disorder, attention- deficit/hyperactivity disorder, stress regulation |
DRD4 |
rs1800443 |
[G/T] |
0.029 |
Behavioral dyscontrol, impulsivity, attention deficit/hyperactivity disorder |
DBH |
rs1611115 |
[C/T] |
0.175 |
Behavioral dyscontrol, mood and anxiety disorders, impulsivity |
SLC6A3 |
Vntr |
10 repeats |
0.71 |
Behavioral dyscontrol, attention deficit/hyperactivity disorder |
CHRNA5 |
rs16969968 |
[A/G] |
0.424 |
Nicotine dependence, alcohol abuse and dependence (cholinergic system) |
*Allele frequencies have been documented for various populations. Those shown are for Caucasians of European ancestry. |
||||
Table 2. Full listing of minor allele frequencies (MAF) for selected genetic markers, for individuals of various ethnic origins |
|
|
|
|
||||||||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
MAF |
MAF |
MAF |
MAF |
MAF |
|
|
|
|
|
|
|
SNP_Name |
Chr |
Coordinate |
Caucasian |
African |
African Amer. |
Japanese |
Chinese |
Gene_ID |
Gene_symbol |
Accession |
|
|
|
|
rs2376805 |
1 |
1946222 |
-99 |
-99 |
-99 |
-99 |
-99 |
2563 |
GABRD |
NM_000815.2 |
|
|
|
|
rs204076 |
1 |
29062977 |
0.358 |
0.133 |
0.2333 |
0.057 |
0.078 |
4985 |
OPRD1 |
NM_000911.3 |
|
|
|
|
rs913168 |
1 |
46631163 |
0.424 |
0.408 |
-99 |
0.25 |
0.389 |
2166 |
FAAH |
NM_001441.1 |
|
|
|
|
rs4845652 |
1 |
152804829 |
0.093 |
0.192 |
-99 |
0.125 |
0.1 |
1141 |
CHRNB2 |
NM_000748.1 |
|
|
|
|
rs3891068 |
1 |
204398689 |
-99 |
-99 |
-99 |
-99 |
-99 |
553 |
AVPR1B |
NM_000707.2 |
|
|
|
|
rs6432224 |
2 |
11718851 |
0 |
0.158 |
-99 |
0 |
0 |
23620 |
NTSR2 |
NM_012344.2 |
|
|
|
|
rs6719226 |
2 |
25249516 |
0.058 |
0.383 |
-99 |
0.239 |
0.078 |
5443 |
POMC |
NM_001035256.1 |
|
|
|
|
rs6709781 |
2 |
27384369 |
-99 |
-99 |
-99 |
-99 |
-99 |
7349 |
UCN |
NM_003353.2 |
|
|
|
|
rs4538251 |
2 |
45739306 |
-99 |
-99 |
-99 |
-99 |
-99 |
5581 |
PRKCE |
NM_005400.2 |
|
|
|
|
rs2111375 |
2 |
75281386 |
-99 |
-99 |
-99 |
-99 |
-99 |
6869 |
TACR1 |
NM_001058.2 |
|
|
|
|
rs2312955 |
2 |
96140895 |
0.292 |
0.1 |
-99 |
0.307 |
0.489 |
151 |
ADRA2B |
NM_000682.4 |
|
|
|
|
rs11898177 |
2 |
119851961 |
0.317 |
0.467 |
-99 |
0.364 |
0.444 |
1622 |
DBI |
NM_020548.4 |
|
|
|
|
rs13402440 |
2 |
138475664 |
0 |
0.017 |
-99 |
0 |
0 |
3176 |
HNMT |
NM_006895.2 |
|
|
|
|
rs1978340 |
2 |
171378367 |
0.308 |
0.067 |
-99 |
0.182 |
0.222 |
2571 |
GAD1 |
NM_000817.2 |
|
|
|
|
rs769399 |
2 |
171425646 |
0 |
0.042 |
-99 |
0 |
0 |
2571 |
GAD1 |
NM_000817.2 |
|
|
|
|
rs2709376 |
2 |
208098633 |
0.05 |
0.35 |
-99 |
0 |
0 |
1385 |
CREB1 |
NM_004379.2 |
|
|
|
|
rs3806545 |
2 |
231698344 |
0.0625 |
0.442 |
-99 |
-99 |
0.2917 |
3357 |
HTR2B |
NM_000867.2 |
|
|
|
|
rs2600072 |
3 |
10832067 |
0.224 |
0.153 |
-99 |
0.489 |
0.478 |
6538 |
SLC6A11 |
NM_014229.1 |
|
|
|
|
rs13069836 |
3 |
42282913 |
0.45 |
0.381 |
-99 |
0.422 |
0.444 |
885 |
CCK |
NM_000729.3 |
|
|
|
|
rs13319651 |
3 |
48574845 |
0.017 |
0.093 |
-99 |
-99 |
0.081 |
90226 |
UCN2 |
NM_033199.3 |
|
|
|
|
rs3732791 |
3 |
115330379 |
0 |
0 |
-99 |
-99 |
0.011 |
1814 |
DRD3 |
NM_000796.3 |
|
|
|
|
rs7678463 |
4 |
3740750 |
0.167 |
0.195 |
-99 |
0.477 |
0.467 |
152 |
ADRA2C |
NM_000683.3 |
|
|
|
|
rs4516717 |
4 |
9385751 |
-99 |
0.183 |
-99 |
0 |
0 |
1816 |
DRD5 |
NM_000798.3 |
|
|
|
|
rs4349588 |
4 |
26101460 |
0 |
0.083 |
-99 |
0 |
0 |
886 |
CCKAR |
NM_000730.2 |
|
|
|
|
rs1504497 |
4 |
45643201 |
0.308 |
0.042 |
0.15 |
0.182 |
0.311 |
2565 |
GABRG1 |
NM_173536.3 |
|
|
|
|
rs11503014 |
4 |
46085622 |
0.1944 |
0.3158 |
-99 |
-99 |
0.0238 |
2555 |
GABRA2 |
NM_000807.1 |
|
|
|
|
rs1159396 |
4 |
46631579 |
0.483 |
0.408 |
-99 |
0.352 |
0.3 |
2557 |
GABRA4 |
NM_000809.2 |
|
|
|
|
rs10028945 |
4 |
47123062 |
0.25 |
0.3261 |
-99 |
-99 |
0.0833 |
2560 |
GABRB1 |
NM_000812.2 |
|
|
|
|
rs1126671 |
4 |
100267437 |
0.333 |
0.1 |
-99 |
0 |
0 |
127 |
ADH4 |
NM_000670.3 |
|
|
|
|
rs1229984 |
4 |
100458342 |
0 |
0 |
-99 |
0.261 |
0.233 |
125 |
ADH1B |
NM_000668.3 |
|
|
|
|
rs1693482 |
4 |
100482988 |
0.484 |
0.0625 |
-99 |
-99 |
-99 |
126 |
ADH1C |
NM_000669.3 |
|
|
|
|
rs1573496 |
4 |
100568692 |
0.092 |
0 |
0 |
0 |
0 |
131 |
ADH7 |
NM_000673.3 |
|
|
|
|
rs10213647 |
4 |
156346608 |
0.406 |
0.148 |
-99 |
0.444 |
0.466 |
4887 |
NPY2R |
NM_000910.2 |
|
|
|
|
rs10517662 |
4 |
158312511 |
0.058 |
0.133 |
-99 |
0 |
0.022 |
2743 |
GLRB |
NM_000824.2 |
|
|
|
|
rs4057797 |
4 |
164464503 |
0.308 |
0.2 |
-99 |
-99 |
0.4348 |
4886 |
NPY1R |
NM_000909.4 |
|
|
|
|
rs6536721 |
4 |
164496347 |
0.358 |
0.308 |
-99 |
0.341 |
0.311 |
4889 |
NPY5R |
NM_006174.2 |
|
|
|
|
rs27072 |
5 |
1447522 |
0.183 |
0.133 |
-99 |
0.133 |
0.378 |
6531 |
SLC6A3 |
NM_001044.2 |
|
|
|
|
rs1364043 |
5 |
63286607 |
0.217 |
0.183 |
0.2 |
0.3 |
0.378 |
3350 |
HTR1A |
NM_000524.2 |
|
|
|
|
rs7731997 |
5 |
71052324 |
0 |
0.075 |
-99 |
0 |
0 |
9607 |
CARTPT |
NM_004291.2 |
|
|
|
|
rs3792738 |
5 |
76283540 |
0.058 |
0.085 |
-99 |
0.133 |
0.178 |
1393 |
CRHBP |
NM_001882.3 |
|
|
|
|
rs10482605 |
5 |
142763714 |
-99 |
-99 |
-99 |
-99 |
-99 |
2908 |
NR3C1 |
NM_001020825.1 |
|
|
|
|
rs12654778 |
5 |
148185934 |
0.336 |
0.203 |
-99 |
0.333 |
0.356 |
154 |
ADRB2 |
NM_000024.3 |
|
|
|
|
rs2240795 |
5 |
149569840 |
0.483 |
0.058 |
-99 |
0.344 |
0.367 |
6534 |
SLC6A7 |
NM_014228.2 |
|
|
|
|
rs2915885 |
5 |
151187261 |
0.408 |
0.449 |
-99 |
0.261 |
0.244 |
2741 |
GLRA1 |
NM_000171.1 |
|
|
|
|
rs4426954 |
5 |
160885609 |
0.407 |
0.175 |
-99 |
0.378 |
0.456 |
2561 |
GABRB2 |
NM_000813.1 |
|
|
|
|
rs3811991 |
5 |
161060987 |
0.3043 |
0.4565 |
-99 |
-99 |
0.1875 |
2559 |
GABRA6 |
NM_000811.1 |
|
|
|
|
rs209345 |
5 |
161423965 |
0.026 |
0.183 |
-99 |
0 |
0.011 |
2566 |
GABRG2 |
NM_198904.1 |
|
|
|
|
rs10078866 |
5 |
174804926 |
0 |
0 |
-99 |
0 |
0 |
1812 |
DRD1 |
NM_000794.3 |
|
|
|
|
rs3800373 |
6 |
35650454 |
0.3125 |
0.4783 |
-99 |
-99 |
0.2917 |
2289 |
FKBP5 |
NM_004117.2 |
|
|
|
|
rs851027 |
6 |
36098853 |
0.292 |
0.217 |
-99 |
0.443 |
0.344 |
116369 |
SLC26A8 |
NM_138718.1 |
|
|
|
|
rs7761118 |
6 |
36176281 |
0.108 |
0.241 |
-99 |
0.105 |
0.178 |
1432 |
MAPK14 |
NM_001315.1 |
|
|
|
|
rs507964 |
6 |
44294368 |
0 |
-99 |
-99 |
-99 |
-99 |
2030 |
SLC29A1 |
NM_004955.1 |
|
|
|
|
rs1213366 |
6 |
78233589 |
0.39 |
0.042 |
-99 |
0.068 |
0.111 |
3351 |
HTR1B |
NM_000863.1 |
|
|
|
|
rs6911472 |
6 |
88909862 |
0 |
0.042 |
-99 |
0 |
0 |
1268 |
CNR1 |
NM_033181.2 |
|
|
|
|
rs7770466 |
6 |
146800958 |
0.025 |
0.242 |
-99 |
0 |
0 |
2911 |
GRM1 |
NM_000838.2 |
|
|
|
|
rs1799972 |
6 |
154402389 |
0.016 |
0.2085 |
-99 |
-99 |
-99 |
4988 |
OPRM1 |
NM_001008504.1 |
|
|
|
|
rs4314511 |
6 |
154607027 |
0.158 |
0.05 |
-99 |
0.364 |
0.307 |
26034 |
PIP3-E |
NM_015553.1 |
|
|
|
|
rs10370 |
6 |
160021522 |
0.47 |
-99 |
-99 |
-99 |
-99 |
6648 |
SOD2 |
NM_001024466.1 |
|
|
|
|
rs16475 |
7 |
24298011 |
0.045 |
0 |
-99 |
-99 |
0.011 |
4852 |
NPY |
NM_000905.2 |
|
|
|
|
rs8192492 |
7 |
30659687 |
0 |
0 |
-99 |
-99 |
0 |
1395 |
CRHR2 |
NM_001883.2 |
|
|
|
|
rs7804365 |
7 |
50604642 |
0.442 |
0.119 |
-99 |
0.302 |
0.33 |
1644 |
DDC |
NM_000790.2 |
|
|
|
|
rs6465606 |
7 |
97192511 |
0.258 |
0.35 |
-99 |
0.08 |
0.1 |
6863 |
TAC1 |
NM_003182.1 |
|
|
|
|
rs3828942 |
7 |
127681541 |
0.492 |
0.108 |
-99 |
0.227 |
0.278 |
3952 |
LEP |
NM_000230.1 |
|
|
|
|
rs10266026 |
7 |
136201512 |
0.008 |
0.075 |
-99 |
0 |
0.011 |
1129 |
CHRM2 |
NM_001006630.1 |
|
|
|
|
rs7820517 |
8 |
20088916 |
0.127 |
0.25 |
-99 |
0.318 |
0.344 |
6570 |
SLC18A1 |
NM_003053.1 |
|
|
|
|
rs2291776 |
8 |
26667553 |
0.068 |
0.158 |
-99 |
0.341 |
0.311 |
148 |
ADRA1A |
NM_033302.1 |
|
|
|
|
rs7823854 |
8 |
27393424 |
0 |
0.188 |
-99 |
-99 |
0 |
1135 |
CHRNA2 |
NM_000742.1 |
|
|
|
|
rs2722897 |
8 |
28229116 |
0.125 |
0.492 |
-99 |
0.256 |
0.133 |
5368 |
PNOC |
NM_006228.3 |
|
|
|
|
rs6989250 |
8 |
54328948 |
0 |
0.125 |
-99 |
0 |
0 |
4986 |
OPRK1 |
NM_000912.3 |
|
|
|
|
rs11998459 |
8 |
57516941 |
0 |
0.033 |
-99 |
0 |
0 |
5179 |
PENK |
NM_006211.2 |
|
|
|
|
rs5030875 |
8 |
67256620 |
0.042 |
0.275 |
-99 |
0 |
0 |
1392 |
CRH |
NM_000756.1 |
|
|
|
|
rs1187321 |
9 |
86472851 |
0.192 |
0.433 |
-99 |
0.239 |
0.233 |
4915 |
NTRK2 |
NM_006180.3 |
|
|
|
|
rs77905 |
9 |
135507918 |
0.458 |
0.383 |
-99 |
0.182 |
0.067 |
1621 |
DBH |
NM_000787.2 |
|
|
|
|
rs2071644 |
9 |
139178730 |
0 |
0 |
-99 |
0.011 |
0.011 |
2902 |
GRIN1 |
NM_000832.4 |
|
|
|
|
rs10904478 |
10 |
5394347 |
0.475 |
0.192 |
-99 |
0.389 |
0.467 |
114131 |
UCN3 |
NM_053049.2 |
|
|
|
|
rs4747550 |
10 |
26625608 |
0.167 |
0.408 |
-99 |
0.43 |
0.389 |
2572 |
GAD2 |
NM_000818.1 |
|
|
|
|
rs638019 |
10 |
112821819 |
0.3542 |
0.067 |
-99 |
0.318 |
0.322 |
150 |
ADRA2A |
NM_000681.2 |
|
|
|
|
rs363276 |
10 |
119023799 |
0.117 |
0.333 |
-99 |
0.318 |
0.422 |
6571 |
SLC18A2 |
NM_003054.2 |
|
|
|
|
rs3813865 |
10 |
135189234 |
0.025 |
0.1053 |
-99 |
-99 |
0.3478 |
1571 |
CYP2E1 |
NM_000773.3 |
|
|
|
|
rs11246226 |
11 |
631191 |
0.408 |
0.379 |
-99 |
0.244 |
0.239 |
1815 |
DRD4 |
NM_000797.2 |
|
|
|
|
rs2070762 |
11 |
2142911 |
0.483 |
0.161 |
-99 |
0.456 |
0.455 |
7054 |
TH |
NM_000360.3 |
|
|
|
|
rs1805002 |
11 |
6247696 |
0.042 |
0.092 |
-99 |
0.068 |
0.011 |
887 |
CCKBR |
NM_176875.2 |
|
|
|
|
rs10741734 |
11 |
18001224 |
0.4 |
0.142 |
-99 |
0.489 |
0.443 |
7166 |
TPH1 |
NM_004179.1 |
|
|
|
|
rs908867 |
11 |
27702340 |
0.117 |
0.1 |
-99 |
0.078 |
0.011 |
627 |
BDNF |
NM_170731.3 |
|
|
|
|
rs4930241 |
11 |
68205159 |
-99 |
-99 |
-99 |
-99 |
-99 |
51083 |
GAL |
NM_015973.3 |
|
|
|
|
rs12364283 |
11 |
112852165 |
0.075 |
0.008 |
-99 |
0 |
0.011 |
1813 |
DRD2 |
NM_000795.2 |
|
|
|
|
rs3758987 |
11 |
113280485 |
0.219 |
0.466 |
-99 |
0.35 |
0.151 |
9177 |
HTR3B |
NM_006028.3 |
|
|
|
|
rs1176713 |
11 |
113365635 |
0.302 |
0.314 |
-99 |
0.273 |
0.211 |
3359 |
HTR3A |
NM_000869.2 |
|
|
|
|
rs570680 |
12 |
200576 |
-99 |
-99 |
-99 |
-99 |
-99 |
6540 |
SLC6A13 |
NM_016615.2 |
|
|
|
|
rs1895056 |
12 |
14029302 |
0.258 |
0 |
-99 |
0 |
0.011 |
2904 |
GRIN2B |
NM_000834.2 |
|
|
|
|
rs3803107 |
12 |
61827101 |
0.142 |
0.325 |
-99 |
0.193 |
0.144 |
552 |
AVPR1A |
NM_000706.3 |
|
|
|
|
rs11179085 |
12 |
70824696 |
0.017 |
0.119 |
-99 |
0.102 |
0.133 |
121278 |
TPH2 |
NM_173353.2 |
|
|
|
|
rs6314 |
13 |
46307035 |
0.075 |
0.158 |
-99 |
0 |
0 |
3356 |
HTR2A |
NM_000621.2 |
|
|
|
|
rs2273836 |
14 |
66716010 |
0.0208 |
0.0217 |
-99 |
-99 |
0 |
10243 |
GPHN |
NM_001024218.1 |
|
|
|
|
rs11853694 |
15 |
24347672 |
0 |
0.4 |
-99 |
0 |
0 |
2562 |
GABRB3 |
NM_000814.4 |
|
|
|
|
rs2376481 |
15 |
24803459 |
0.45 |
0.325 |
-99 |
0.489 |
0.456 |
2558 |
GABRA5 |
NM_000810.2 |
|
|
|
|
rs140672 |
15 |
25154938 |
0 |
0.3478 |
-99 |
-99 |
0.0417 |
2567 |
GABRG3 |
NM_033223.1 |
|
|
|
|
rs6494223 |
15 |
30183749 |
0.433 |
0.492 |
-99 |
0.344 |
0.433 |
1139 |
CHRNA7 |
NM_000746.3 |
|
|
|
|
rs684513 |
15 |
76645455 |
0.217 |
0.158 |
-99 |
0.239 |
0.278 |
1138 |
CHRNA5 |
NM_000745.2 |
|
|
|
|
rs1878399 |
15 |
76699058 |
0.375 |
0.225 |
-99 |
0.159 |
0.178 |
1136 |
CHRNA3 |
NM_000743.2 |
|
|
|
|
rs6495309 |
15 |
76702300 |
0.2 |
0.25 |
-99 |
-99 |
-99 |
1143 |
CHRNB4 |
NM_000750.2 |
|
|
|
|
rs1071504 |
16 |
10187517 |
0.358 |
0.408 |
-99 |
0.1 |
0.1 |
2903 |
GRIN2A |
NM_000833.2 |
|
|
|
|
rs7698 |
16 |
30033301 |
0.125 |
-99 |
-99 |
-99 |
-99 |
5595 |
MAPK3 |
NM_002746.2 |
|
|
|
|
rs1064448 |
16 |
48908384 |
0.492 |
0.058 |
-99 |
0.2 |
0.1 |
113 |
ADCY7 |
NM_001114.2 |
|
|
|
|
rs1532703 |
16 |
54243985 |
0.017 |
0.017 |
-99 |
0 |
0 |
6530 |
SLC6A2 |
NM_001043.2 |
|
|
|
|
rs4522461 |
17 |
4568522 |
0.217 |
0.117 |
-99 |
0 |
0 |
409 |
ARRB2 |
NM_004313.3 |
|
|
|
|
rs3813034 |
17 |
25548930 |
-99 |
-99 |
-99 |
-99 |
-99 |
6532 |
SLC6A4 |
NM_001045.2 |
|
|
|
|
rs734645 |
17 |
35045819 |
0.0417 |
0.25 |
-99 |
-99 |
0 |
84152 |
PPP1R1B |
NM_032192.2 |
|
|
|
|
rs2293153 |
17 |
37581569 |
0 |
0.458 |
-99 |
0.034 |
0.011 |
23415 |
KCNH4 |
NM_012285.1 |
|
|
|
|
rs7226205 |
17 |
37594481 |
0 |
0.008 |
-99 |
0 |
0 |
84514 |
GHDC |
NM_032484.3 |
|
|
|
|
rs9900679 |
17 |
41223923 |
0 |
0.1 |
-99 |
0 |
0 |
1394 |
CRHR1 |
NM_004382.2 |
|
|
|
|
rs2683267 |
17 |
70366578 |
0.225 |
0.492 |
-99 |
0.398 |
0.467 |
2905 |
GRIN2C |
NM_000835.3 |
|
|
|
|
rs9916013 |
17 |
71583394 |
0 |
0.033 |
-99 |
0 |
0 |
8811 |
GALR2 |
NM_003857.2 |
|
|
|
|
rs949060 |
18 |
73087926 |
0.442 |
0.1 |
-99 |
0.205 |
0.2 |
2587 |
GALR1 |
NM_001480.2 |
|
|
|
|
rs6136667 |
20 |
1926301 |
0.127 |
0 |
-99 |
0.463 |
0.317 |
5173 |
PDYN |
NM_024411.2 |
|
|
|
|
rs4813625 |
20 |
2997720 |
0.467 |
0.491 |
-99 |
0.455 |
0.444 |
5020 |
OXT |
NM_000915.2 |
|
|
|
|
rs1321099 |
20 |
36792059 |
0.058 |
0.4 |
-99 |
0.455 |
0.467 |
140679 |
SLC32A1 |
NM_080552.2 |
|
|
|
|
rs2427400 |
20 |
60806120 |
0.125 |
0.1 |
-99 |
0.163 |
0.133 |
4923 |
NTSR1 |
NM_002531.1 |
|
|
|
|
rs755203 |
20 |
61464708 |
0.383 |
0.033 |
0.0909 |
0.442 |
0.344 |
1137 |
CHRNA4 |
NM_000744.3 |
|
|
|
|
rs6090041 |
20 |
62183120 |
0.3 |
0.442 |
-99 |
0.093 |
0.067 |
4987 |
OPRL1 |
NM_182647.1 |
|
|
|
|
rs2832489 |
21 |
30221341 |
0.233 |
0.4 |
-99 |
0.148 |
0.144 |
2897 |
GRIK1 |
NM_175611.2 |
|
|
|
|
rs737866 |
22 |
18310109 |
0.317 |
0.142 |
-99 |
0.352 |
0.322 |
10587 |
TXNRD2 |
NM_006440.3 |
|
|
|
|
rs9332377 |
22 |
18335692 |
0.167 |
0.375 |
-99 |
0 |
0 |
1312 |
COMT |
NM_007310.1 |
|
|
|
|
rs5751862 |
22 |
23132564 |
0.5 |
0.117 |
-99 |
0.432 |
0.478 |
23384 |
SPECC1L |
NM_015330.1 |
|
|
|
|
rs4822492 |
22 |
23173594 |
0.4 |
0.175 |
-99 |
0.42 |
0.467 |
135 |
ADORA2A |
NM_000675.3 |
|
|
|
|
rs4821717 |
22 |
36546704 |
0.367 |
0.058 |
-99 |
0.216 |
0.078 |
8484 |
GALR3 |
NM_003614.1 |
|
|
|
|
rs3091367 |
22 |
36557767 |
0.175 |
0.3261 |
-99 |
-99 |
0.0833 |
129138 |
ANKRD54 |
NM_138797.1 |
|
|
|
|
rs6001728 |
22 |
38764703 |
0.233 |
0.067 |
-99 |
0.034 |
0.033 |
23112 |
TNRC6B |
NM_001024843.1 |
|
|
|
|
rs1033372 |
22 |
48249853 |
0.008 |
0.133 |
-99 |
0.395 |
0.456 |
728652 |
LOC728652 |
XM_001128024.1 |
|
|
|
|
rs6526714 |
X |
14485073 |
0.4444 |
0.2111 |
-99 |
0.1642 |
0.2647 |
2742 |
GLRA2 |
NM_002063.2 |
|
|
|
|
rs2239448 |
X |
43487623 |
0.2333 |
0.4 |
-99 |
0.3788 |
0.3333 |
4128 |
MAOA |
NM_000240.2 |
|
|
|
|
rs1799836 |
X |
43512943 |
0.3833 |
0.2333 |
-99 |
0.1591 |
0.1304 |
4129 |
MAOB |
NM_000898.3 |
|
|
|
|
rs508865 |
X |
113720371 |
0.4 |
0.3833 |
-99 |
0.2045 |
0.1304 |
3358 |
HTR2C |
NM_000868.1 |
|
|
|
|
rs1061421 |
X |
150873252 |
0.0833 |
0.45 |
-99 |
0 |
0.0217 |
2564 |
GABRE |
NM_004961.3 |
|
|
|
|
rs5970292 |
X |
151311524 |
0.4 |
0.2889 |
-99 |
0.2424 |
0.2424 |
2556 |
GABRA3 |
NM_000808.2 |
|
|
|
|
rs3810651 |
X |
151571933 |
0.4111 |
0.314 |
-99 |
0.2537 |
0.3824 |
55879 |
GABRQ |
NM_018558.1 |
|
|
|
|
rs6670693 |
1 |
1095975 |
0 |
0.025 |
-99 |
0 |
0 |
254173 |
TTLL10 |
NM_153254.1 |
|
|
|
|
rs3100865 |
1 |
2795967 |
0.267 |
0.008 |
-99 |
0.409 |
0.278 |
728702 |
LOC728702 |
XM_001128239.1 |
|
|
|
|
rs4653130 |
1 |
35576728 |
0.067 |
0.392 |
-99 |
0.284 |
0.144 |
9202 |
ZMYM4 |
NM_005095.2 |
|
|
|
|
rs655590 |
1 |
35894634 |
0.058 |
0.325 |
-99 |
0.295 |
0.111 |
5690 |
PSMB2 |
NM_002794.3 |
|
|
|
|
rs6687440 |
1 |
106571633 |
0.3 |
0.083 |
-99 |
0.011 |
0.022 |
126987 |
LOC126987 |
XR_016153.1 |
|
|
|
|
rs11184898 |
1 |
106755851 |
0.442 |
0 |
-99 |
0.045 |
0.044 |
126987 |
LOC126987 |
XR_016153.1 |
|
|
|
|
rs10802184 |
1 |
116680864 |
0.1 |
0.017 |
-99 |
0.068 |
0.022 |
476 |
ATP1A1 |
NM_000701.6 |
|
|
|
|
rs1446959 |
1 |
157873428 |
0.433 |
0.042 |
-99 |
0.068 |
0.078 |
325 |
APCS |
NM_001639.2 |
|
|
|
|
rs4265409 |
1 |
161681183 |
0.392 |
0.133 |
-99 |
0.023 |
0.044 |
83540 |
NUF2 |
NM_145697.1 |
|
|
|
|
rs6424922 |
1 |
182240761 |
0.158 |
0.05 |
-99 |
0.33 |
0.333 |
23127 |
GLT25D2 |
NM_015101.2 |
|
|
|
|
rs2220128 |
1 |
184002536 |
0.267 |
0.075 |
-99 |
0 |
0.022 |
83872 |
HMCN1 |
NM_031935.1 |
|
|
|
|
rs2759281 |
1 |
203132988 |
0.15 |
0.017 |
-99 |
0.2 |
0.122 |
23114 |
NFASC |
NM_015090.2 |
|
|
|
|
rs6718709 |
2 |
5257953 |
0.45 |
0.175 |
-99 |
0.102 |
0.022 |
6664 |
SOX11 |
NM_003108.3 |
|
|
|
|
rs1368928 |
2 |
8668914 |
0.342 |
0.042 |
-99 |
0.034 |
0.056 |
3398 |
ID2 |
NM_002166.4 |
|
|
|
|
rs11126303 |
2 |
26027007 |
0.008 |
0.192 |
-99 |
0.091 |
0.078 |
3797 |
KIF3C |
NM_002254.5 |
|
|
|
|
rs12618959 |
2 |
33017154 |
0.308 |
0.092 |
-99 |
0.023 |
0.044 |
4052 |
LTBP1 |
NM_206943.1 |
|
|
|
|
rs1811510 |
3 |
5750231 |
0.026 |
0.05 |
-99 |
0.205 |
0.167 |
9695 |
EDEM1 |
NM_014674.1 |
|
|
|
|
rs6785846 |
3 |
25714245 |
0.208 |
0.042 |
-99 |
0.159 |
0.067 |
55768 |
NGLY1 |
NM_018297.2 |
|
|
|
|
rs1028040 |
3 |
57215650 |
0.392 |
0.1 |
-99 |
0.114 |
0.078 |
8820 |
HESX1 |
NM_003865.1 |
|
|
|
|
rs1563382 |
3 |
79319581 |
0.05 |
0.075 |
-99 |
0 |
0.011 |
6091 |
ROBO1 |
NM_002941.2 |
|
|
|
|
rs6790692 |
3 |
98695905 |
0.183 |
0.042 |
-99 |
0.216 |
0.067 |
285220 |
EPHA6 |
XM_114973.6 |
|
|
|
|
rs1503079 |
3 |
106241722 |
0.492 |
0.133 |
-99 |
0.08 |
0.067 |
214 |
ALCAM |
NM_001627.2 |
|
|
|
|
rs10933921 |
3 |
109082643 |
0.483 |
0.017 |
-99 |
0.341 |
0.089 |
285205 |
LOC285205 |
XM_001128618.1 |
|
|
|
|
rs4241398 |
3 |
110299018 |
0.475 |
0.042 |
-99 |
0.068 |
0.056 |
27136 |
MORC1 |
NM_014429.2 |
|
|
|
|
rs11713100 |
3 |
111513795 |
0.05 |
0.208 |
-99 |
0.307 |
0.478 |
389141 |
LOC389141 |
XR_016945.1 |
|
|
|
|
rs6414248 |
3 |
112365016 |
0.167 |
0.008 |
-99 |
0.17 |
0.322 |
25945 |
PVRL3 |
NM_015480.1 |
|
|
|
|
rs1698042 |
3 |
118667838 |
0.058 |
0.2 |
-99 |
0.489 |
0.478 |
728873 |
LOC728873 |
XR_015679.1 |
|
|
|
|
rs2626018 |
3 |
124774156 |
0.208 |
0.05 |
-99 |
0.352 |
0.3 |
201562 |
PTPLB |
NM_198402.2 |
|
|
|
|
rs2625956 |
3 |
130747960 |
0.142 |
0.033 |
-99 |
0.409 |
0.389 |
132243 |
H1FOO |
NM_153833.1 |
|
|
|
|
rs36110 |
3 |
136182877 |
0.45 |
0.058 |
-99 |
0.034 |
0.111 |
2047 |
EPHB1 |
NM_004441.3 |
|
|
|
|
rs1519260 |
3 |
137062756 |
0.425 |
0.017 |
-99 |
0.216 |
0.122 |
5523 |
PPP2R3A |
NM_002718.3 |
|
|
|
|
rs7618370 |
3 |
169391997 |
0.167 |
0.042 |
-99 |
0.011 |
0.067 |
389174 |
LOC389174 |
XM_371678.4 |
|
|
|
|
rs3796285 |
3 |
189735795 |
0.467 |
0.108 |
-99 |
0.102 |
0.067 |
4026 |
LPP |
NM_005578.2 |
|
|
|
|
rs4687002 |
3 |
190104562 |
0.317 |
0.058 |
-99 |
0.023 |
0.056 |
4026 |
LPP |
NM_005578.2 |
|
|
|
|
rs733370 |
4 |
6010036 |
0.342 |
0.075 |
-99 |
0.216 |
0.111 |
389197 |
FLJ46481 |
NM_207405.1 |
|
|
|
|
rs16877243 |
4 |
25177016 |
0.183 |
0.033 |
-99 |
0.375 |
0.378 |
645433 |
LOC645433 |
XR_017242.1 |
|
|
|
|
rs1488299 |
4 |
26870541 |
0.483 |
0.15 |
-99 |
0.034 |
0.033 |
401123 |
FLJ45721 |
NM_207490.1 |
|
|
|
|
rs1507086 |
4 |
41651170 |
0.3 |
0 |
-99 |
0.023 |
0.022 |
55161 |
TMEM33 |
NM_018126.1 |
|
|
|
|
rs11098964 |
4 |
81106993 |
0.483 |
0.1 |
-99 |
0.023 |
0.044 |
118429 |
ANTXR2 |
NM_058172.3 |
|
|
|
|
rs1444893 |
4 |
82602473 |
0.208 |
0.067 |
-99 |
0.023 |
0.022 |
153020 |
RASGEF1B |
NM_152545.1 |
|
|
|
|
rs1494962 |
4 |
84555307 |
0.117 |
0.092 |
-99 |
0 |
0.011 |
113510 |
HEL308 |
NM_133636.1 |
|
|
|
|
rs2231164 |
4 |
89234881 |
-99 |
-99 |
-99 |
-99 |
-99 |
9429 |
ABCG2 |
NM_004827.2 |
|
|
|
|
rs13134663 |
4 |
110060008 |
0.492 |
0.108 |
-99 |
0.068 |
0.067 |
84570 |
COL25A1 |
NM_032518.2 |
|
|
|
|
rs1827950 |
4 |
117317931 |
0.083 |
0.033 |
-99 |
0.08 |
0.078 |
645368 |
LOC645368 |
XR_017440.1 |
|
|
|
|
rs4863731 |
4 |
141184618 |
0.433 |
0.05 |
-99 |
0.034 |
0.056 |
55534 |
MAML3 |
NM_018717.3 |
|
|
|
|
rs1397529 |
4 |
144611612 |
0.367 |
0.042 |
-99 |
0.045 |
0.044 |
2549 |
GAB1 |
NM_207123.1 |
|
|
|
|
rs7692206 |
4 |
155326143 |
0.083 |
0.033 |
-99 |
0.341 |
0.256 |
54798 |
DCHS2 |
NM_017639.2 |
|
|
|
|
rs2345275 |
4 |
158944273 |
0.483 |
0.083 |
-99 |
0.089 |
0.111 |
729937 |
LOC729937 |
XM_001131838.1 |
|
|
|
|
rs12644851 |
4 |
159827612 |
0.383 |
0.008 |
-99 |
0.216 |
0.078 |
2110 |
ETFDH |
NM_004453.1 |
|
|
|
|
rs6552216 |
4 |
178348998 |
0.192 |
0.358 |
-99 |
0.125 |
0.078 |
55247 |
NEIL3 |
NM_018248.1 |
|
|
|
|
rs26880 |
5 |
4958892 |
0.425 |
0.075 |
-99 |
0.189 |
0.122 |
340094 |
LOC340094 |
XM_295155.5 |
|
|
|
|
rs246760 |
5 |
5633216 |
0.325 |
0.05 |
-99 |
0.278 |
0.156 |
23379 |
KIAA0947 |
XM_029101.11 |
|
|
|
|
rs2937067 |
5 |
116167937 |
0.425 |
0.05 |
-99 |
0.078 |
0.133 |
57556 |
SEMA6A |
NM_020796.2 |
|
|
|
|
rs2220858 |
5 |
116660106 |
0.367 |
0.05 |
-99 |
0.068 |
0.067 |
728342 |
LOC728342 |
XM_001129097.1 |
|
|
|
|
rs267071 |
5 |
116976402 |
0.319 |
0.025 |
-99 |
0.034 |
0.022 |
728342 |
LOC728342 |
XM_001129097.1 |
|
|
|
|
rs2416504 |
5 |
117436991 |
0.405 |
0.008 |
-99 |
0.023 |
0.022 |
728342 |
LOC728342 |
XM_001129097.1 |
|
|
|
|
rs6595142 |
5 |
117895090 |
0.367 |
0.033 |
-99 |
0.122 |
0.144 |
285605 |
DTWD2 |
NM_173666.1 |
|
|
|
|
rs2036565 |
5 |
118560770 |
0.442 |
0.175 |
-99 |
0.045 |
0.033 |
1657 |
DMXL1 |
NM_005509.3 |
|
|
|
|
rs326626 |
5 |
133639706 |
0.042 |
0.275 |
-99 |
0.056 |
0.067 |
51265 |
CDKL3 |
NM_016508.2 |
|
|
|
|
rs4958667 |
5 |
152940571 |
0.383 |
0.042 |
-99 |
0.244 |
0.067 |
2890 |
GRIA1 |
NM_000827.2 |
|
|
|
|
rs815608 |
5 |
153496967 |
0.367 |
0.046 |
-99 |
0.027 |
0.056 |
55568 |
GALNT10 |
NM_198321.2 |
|
|
|
|
rs692713 |
5 |
176186041 |
0.042 |
0.042 |
-99 |
0.08 |
0.122 |
90249 |
UNC5A |
NM_133369.2 |
|
|
|
|
rs6869589 |
5 |
178559215 |
0 |
0.017 |
-99 |
0 |
0 |
9509 |
ADAMTS2 |
NM_021599.1 |
|
|
|
|
rs6926774 |
6 |
5677819 |
0.417 |
0.042 |
-99 |
0.102 |
0.078 |
10667 |
FARS2 |
NM_006567.2 |
|
|
|
|
rs1206920 |
6 |
9867494 |
0.308 |
0.167 |
-99 |
0.08 |
0.044 |
266553 |
OFCC1 |
NM_153003.1 |
|
|
|
|
rs2842063 |
6 |
71295098 |
0.183 |
0.05 |
-99 |
0.17 |
0.056 |
57579 |
FAM135A |
NM_020819.2 |
|
|
|
|
rs1538956 |
6 |
127005719 |
0.425 |
0.15 |
-99 |
0 |
0.011 |
728666 |
LOC728666 |
XR_015645.1 |
|
|
|
|
rs9388989 |
6 |
132739275 |
0.117 |
0.017 |
-99 |
0.267 |
0.244 |
26002 |
MOXD1 |
NM_001031699.1 |
|
|
|
|
rs1052502 |
6 |
135648258 |
0.05 |
0.058 |
-99 |
0.136 |
0.144 |
54806 |
AHI1 |
NM_017651.3 |
|
|
|
|
rs1554817 |
6 |
154608113 |
0.158 |
0.058 |
-99 |
0.364 |
0.311 |
26034 |
PIP3-E |
NM_015553.1 |
|
|
|
|
rs9639213 |
7 |
14707372 |
0.467 |
0.05 |
-99 |
0.114 |
0.067 |
1607 |
DGKB |
NM_145695.1 |
|
|
|
|
rs4721415 |
7 |
15202657 |
0.058 |
0.358 |
-99 |
0 |
0.011 |
392636 |
TMEM195 |
NM_001004320.1 |
|
|
|
|
rs6955490 |
7 |
98812524 |
0.133 |
0.067 |
-99 |
0.023 |
0.022 |
10095 |
ARPC1B |
NM_005720.2 |
|
|
|
|
rs10249419 |
7 |
117169042 |
0.383 |
0.083 |
-99 |
0.011 |
0 |
83992 |
CTTNBP2 |
NM_033427.2 |
|
|
|
|
rs10488401 |
7 |
135476064 |
0.433 |
0.008 |
-99 |
0.193 |
0.056 |
730221 |
LOC730221 |
XM_001133013.1 |
|
|
|
|
rs10488619 |
7 |
135824107 |
0.408 |
-99 |
-99 |
0.182 |
0.056 |
392100 |
LOC392100 |
XR_017485.1 |
|
|
|
|
rs315280 |
7 |
142552081 |
0.242 |
0.017 |
0.1446 |
0.091 |
0.167 |
5304 |
PIP |
NM_002652.2 |
|
|
|
|
rs11136793 |
8 |
4767247 |
0.45 |
0.075 |
-99 |
0.193 |
0.078 |
64478 |
CSMD1 |
NM_033225.3 |
|
|
|
|
rs4841401 |
8 |
10527002 |
0.358 |
0 |
-99 |
0.102 |
0.089 |
94137 |
RP1L1 |
NM_178857.4 |
|
|
|
|
rs10107384 |
8 |
10890860 |
0.35 |
0.008 |
-99 |
0.057 |
0.033 |
286046 |
XKR6 |
NM_173683.3 |
|
|
|
|
rs2409710 |
8 |
11017231 |
0.475 |
0.042 |
-99 |
0.068 |
0.056 |
439940 |
C8orf15 |
NM_001033662.1 |
|
|
|
|
rs1347201 |
8 |
16581855 |
0.458 |
0.125 |
-99 |
0.057 |
0.033 |
26281 |
FGF20 |
NM_019851.1 |
|
|
|
|
rs12678324 |
8 |
16833064 |
0.058 |
0.067 |
-99 |
0.466 |
0.5 |
26281 |
FGF20 |
NM_019851.1 |
|
|
|
|
rs2927385 |
8 |
20744320 |
0.45 |
0.042 |
-99 |
0.227 |
0.111 |
646608 |
LOC646608 |
XM_929545.1 |
|
|
|
|
rs2102727 |
8 |
53063166 |
0.158 |
0.008 |
-99 |
0.341 |
0.344 |
9705 |
ST18 |
NM_014682.1 |
|
|
|
|
rs4737761 |
8 |
54867106 |
0.083 |
0.008 |
-99 |
0.466 |
0.4 |
51606 |
ATP6V1H |
NM_213619.1 |
|
|
|
|
rs3912537 |
8 |
62354583 |
0.108 |
0.475 |
-99 |
0.189 |
0.089 |
157807 |
RLBP1L1 |
NM_173519.1 |
|
|
|
|
rs6991838 |
8 |
66633516 |
0.442 |
0.05 |
-99 |
0.102 |
0.1 |
55156 |
ARMC1 |
NM_018120.3 |
|
|
|
|
rs6472362 |
8 |
68994970 |
0.225 |
0.067 |
-99 |
0.068 |
0.022 |
80243 |
DEPDC2 |
NM_025170.4 |
|
|
|
|
rs1227647 |
8 |
80291691 |
0.25 |
0.033 |
-99 |
0.182 |
0.333 |
11075 |
STMN2 |
NM_007029.2 |
|
|
|
|
rs1402851 |
8 |
84417952 |
0.058 |
0.142 |
-99 |
0.284 |
0.5 |
389674 |
HNRPA1P4 |
XM_372050.5 |
|
|
|
|
rs1552314 |
8 |
93173981 |
0.308 |
0.05 |
-99 |
0 |
0.044 |
862 |
RUNX1T1 |
NM_175635.1 |
|
|
|
|
rs4484738 |
8 |
121772608 |
0.25 |
0.017 |
-99 |
0.318 |
0.278 |
6641 |
SNTB1 |
NM_021021.2 |
|
|
|
|
rs1514626 |
8 |
132834923 |
0.2 |
0.058 |
-99 |
0.273 |
0.367 |
23167 |
EFR3A |
NM_015137.1 |
|
|
|
|
rs10113320 |
8 |
134925795 |
0.292 |
0.008 |
-99 |
0.136 |
0.167 |
6482 |
ST3GAL1 |
NM_173344.1 |
|
|
|
|
rs4907376 |
8 |
142602383 |
0.017 |
0.125 |
-99 |
0.465 |
0.489 |
389690 |
FLJ43860 |
NM_207414.1 |
|
|
|
|
rs1871534 |
8 |
145610489 |
0 |
0.017 |
-99 |
0 |
0 |
55630 |
SLC39A4 |
NM_130849.1 |
|
|
|
|
rs2225979 |
9 |
9720381 |
0.475 |
0.05 |
-99 |
0.125 |
0.067 |
5789 |
PTPRD |
NM_002839.1 |
|
|
|
|
rs1885167 |
9 |
17504515 |
0.217 |
0.008 |
-99 |
0.045 |
0.033 |
54875 |
C9orf39 |
NM_017738.1 |
|
|
|
|
rs541805 |
9 |
25924932 |
0.108 |
0.075 |
-99 |
0.489 |
0.478 |
441390 |
-99 |
NM_001004352.1 |
|
|
|
|
rs2486448 |
9 |
71028805 |
0.342 |
0.017 |
-99 |
0.193 |
0.078 |
9414 |
TJP2 |
NM_004817.2 |
|
|
|
|
rs10868793 |
9 |
90352033 |
0.117 |
0.008 |
-99 |
0.17 |
0.2 |
158046 |
NXNL2 |
NM_145283.1 |
|
|
|
|
rs556399 |
9 |
109995109 |
0.475 |
0.125 |
-99 |
0.091 |
0.078 |
392382 |
LOC392382 |
XR_016755.1 |
|
|
|
|
rs2004426 |
9 |
111651570 |
0 |
0.417 |
-99 |
0.136 |
0.089 |
445815 |
PALM2-AKAP2 |
NM_147150.1 |
|
|
|
|
rs2241083 |
9 |
124854714 |
0.167 |
0.025 |
-99 |
0.159 |
0.233 |
23637 |
RABGAP1 |
NM_012197.2 |
|
|
|
|
rs3814134 |
9 |
126307510 |
0.017 |
0.033 |
-99 |
0.182 |
0.311 |
2516 |
NR5A1 |
NM_004959.3 |
|
|
|
|
rs590614 |
9 |
135625230 |
0.392 |
0.033 |
-99 |
0.023 |
0.033 |
7410 |
VAV2 |
NM_003371.2 |
|
|
|
|
rs2388511 |
10 |
3038498 |
0.092 |
0.05 |
-99 |
0.091 |
0.044 |
441546 |
LOC441546 |
XM_499190.2 |
|
|
|
|
rs10795588 |
10 |
8126176 |
0.475 |
0.167 |
-99 |
0.105 |
0.044 |
399717 |
FLJ45983 |
NM_207423.1 |
|
|
|
|
rs2008592 |
10 |
30542827 |
0.083 |
0.167 |
-99 |
0.5 |
0.422 |
57608 |
KIAA1462 |
XM_166132.7 |
|
|
|
|
rs1869237 |
10 |
33876047 |
0.375 |
0.167 |
-99 |
0 |
0.011 |
8829 |
NRP1 |
NM_003873.3 |
|
|
|
|
rs6593430 |
10 |
44422502 |
0.075 |
0 |
-99 |
0 |
0 |
728374 |
LOC728374 |
XR_015274.1 |
|
|
|
|
rs10745288 |
10 |
49801403 |
0.375 |
0.1 |
-99 |
0.023 |
0.022 |
474354 |
LRRC18 |
NM_001006939.2 |
|
|
|
|
rs10763013 |
10 |
55283129 |
0.233 |
0.2 |
-99 |
0 |
0.033 |
65217 |
PCDH15 |
NM_033056.2 |
|
|
|
|
rs11203006 |
10 |
90906065 |
0.15 |
0.483 |
-99 |
0.216 |
0.056 |
728303 |
LOC728303 |
XM_001127130.1 |
|
|
|
|
rs10785952 |
10 |
92038772 |
0.358 |
0.05 |
-99 |
0.011 |
0.033 |
119358 |
LOC119358 |
XM_061427.3 |
|
|
|
|
rs10883533 |
10 |
102462646 |
0.325 |
0.15 |
-99 |
0.034 |
0.033 |
5076 |
PAX2 |
NM_003987.2 |
|
|
|
|
rs10884188 |
10 |
107333364 |
0.392 |
0.008 |
-99 |
0.25 |
0.089 |
22986 |
SORCS3 |
NM_014978.1 |
|
|
|
|
rs1336978 |
10 |
108849094 |
0.408 |
0.025 |
-99 |
0.227 |
0.167 |
114815 |
SORCS1 |
NM_001013031.1 |
|
|
|
|
rs2082380 |
10 |
118144749 |
-99 |
-99 |
-99 |
-99 |
-99 |
374355 |
C10orf96 |
NM_198515.1 |
|
|
|
|
rs1986420 |
10 |
119746836 |
0.017 |
0.083 |
-99 |
0.193 |
0.211 |
22841 |
RAB11FIP2 |
NM_014904.1 |
|
|
|
|
rs2930125 |
10 |
127879147 |
0.15 |
0.058 |
-99 |
0.182 |
0.222 |
8038 |
ADAM12 |
NM_021641.2 |
|
|
|
|
rs10750836 |
11 |
68572099 |
0.008 |
0.217 |
-99 |
0.42 |
0.456 |
219931 |
TPCN2 |
NM_139075.1 |
|
|
|
|
rs2027760 |
11 |
72714129 |
0.1 |
0.333 |
-99 |
0.023 |
0.056 |
9828 |
ARHGEF17 |
NM_014786.2 |
|
|
|
|
rs7932809 |
11 |
121171910 |
0.433 |
0.033 |
-99 |
0.216 |
0.056 |
6653 |
SORL1 |
NM_003105.3 |
|
|
|
|
rs2416791 |
12 |
11592755 |
0.083 |
0.025 |
-99 |
0.284 |
0.278 |
2120 |
ETV6 |
NM_001987.3 |
|
|
|
|
rs2730891 |
12 |
37270952 |
0.042 |
0.075 |
-99 |
0.455 |
0.478 |
144402 |
CPNE8 |
NM_153634.2 |
|
|
|
|
rs1532052 |
12 |
53647581 |
0.233 |
0.342 |
-99 |
0.047 |
0.033 |
9840 |
KIAA0748 |
XM_374983.4 |
|
|
|
|
rs10879311 |
12 |
70352255 |
0.208 |
0.058 |
-99 |
0.125 |
0.211 |
83591 |
THAP2 |
NM_031435.1 |
|
|
|
|
rs711159 |
12 |
78632547 |
0.008 |
0.067 |
-99 |
0.136 |
0.222 |
5074 |
PAWR |
NM_002583.2 |
|
|
|
|
rs12313915 |
12 |
83786783 |
0.308 |
0.05 |
-99 |
0.023 |
0.022 |
55117 |
SLC6A15 |
NM_182767.3 |
|
|
|
|
rs1001484 |
12 |
110021205 |
0.183 |
0.008 |
-99 |
0.33 |
0.333 |
23316 |
CUX2 |
NM_015267.1 |
|
|
|
|
rs628825 |
12 |
110436233 |
0.225 |
0.2 |
0.15 |
0.182 |
0.044 |
6311 |
ATXN2 |
NM_002973.2 |
|
|
|
|
rs1716167 |
12 |
122217115 |
0.2 |
0.05 |
-99 |
0.023 |
0.033 |
10198 |
MPHOSPH9 |
NM_022782.2 |
|
|
|
|
rs10847171 |
12 |
125546608 |
0.267 |
0.042 |
-99 |
0.102 |
0.033 |
728173 |
LOC728173 |
XM_001129061.1 |
|
|
|
|
rs9318026 |
13 |
71204246 |
0.083 |
0.092 |
-99 |
0.205 |
0.2 |
1602 |
DACH1 |
NM_080760.3 |
|
|
|
|
rs4981115 |
14 |
31240826 |
0.125 |
0.092 |
-99 |
0 |
0.011 |
80224 |
NUBPL |
NM_025152.1 |
|
|
|
|
rs7158302 |
14 |
56720501 |
0.083 |
0.1 |
-99 |
0.011 |
0 |
10640 |
EXOC5 |
NM_006544.3 |
|
|
|
|
rs2193595 |
14 |
76914874 |
0.483 |
0.092 |
-99 |
0.091 |
0.067 |
161394 |
C14orf174 |
NM_001010860.1 |
|
|
|
|
rs1003229 |
14 |
97241370 |
0.183 |
0.067 |
-99 |
0.178 |
0.056 |
730217 |
LOC730217 |
XM_001132986.1 |
|
|
|
|
rs1375164 |
15 |
25965407 |
0.175 |
0.008 |
-99 |
0.102 |
0.067 |
4948 |
OCA2 |
NM_000275.1 |
|
|
|
|
rs1828774 |
15 |
26812205 |
0.367 |
0.05 |
-99 |
0.156 |
0.133 |
440253 |
WHDC1L2 |
XM_926785.2 |
|
|
|
|
rs2948905 |
15 |
42933056 |
0.052 |
0.025 |
-99 |
0.455 |
0.378 |
653381 |
LOC653381 |
XR_017364.1 |
|
|
|
|
rs10152524 |
15 |
44027656 |
0.196 |
0.086 |
-99 |
0.011 |
0.013 |
58472 |
SQRDL |
NM_021199.2 |
|
|
|
|
rs2934193 |
15 |
46047011 |
0.092 |
0.025 |
-99 |
0.133 |
0.222 |
283652 |
SLC24A5 |
NM_205850.2 |
|
|
|
|
rs12595448 |
15 |
53691382 |
0.25 |
0 |
-99 |
0.057 |
0.022 |
283659 |
PRTG |
NM_173814.3 |
|
|
|
|
rs289816 |
15 |
61582014 |
0.05 |
0.125 |
-99 |
0.1 |
0.067 |
9960 |
USP3 |
NM_006537.2 |
|
|
|
|
rs10152453 |
15 |
61885666 |
0.058 |
0.192 |
-99 |
0.1 |
0.067 |
8925 |
HERC1 |
NM_003922.2 |
|
|
|
|
rs4511483 |
15 |
69925644 |
0.198 |
0.042 |
-99 |
0.364 |
0.411 |
4649 |
MYO9A |
NM_006901.1 |
|
|
|
|
rs6496858 |
15 |
90062853 |
0.15 |
0.017 |
-99 |
0.5 |
0.411 |
28232 |
SLCO3A1 |
NM_013272.2 |
|
|
|
|
rs1002587 |
16 |
13813024 |
0.2273 |
0.1136 |
-99 |
-99 |
0.1591 |
2072 |
ERCC4 |
NM_005236.1 |
|
|
|
|
rs4783432 |
16 |
21858916 |
0.183 |
0.025 |
-99 |
0.25 |
0.244 |
342293 |
LOC342293 |
XM_292468.4 |
|
|
|
|
rs4787645 |
16 |
30364851 |
0.358 |
0 |
-99 |
0.111 |
0.089 |
22928 |
SEPHS2 |
NM_012248.2 |
|
|
|
|
rs4889490 |
16 |
30730548 |
0.431 |
0 |
-99 |
0.091 |
0.067 |
647086 |
LOC647086 |
XR_017567.1 |
|
|
|
|
rs889548 |
16 |
31045213 |
0.392 |
0.042 |
-99 |
0.111 |
0.056 |
84148 |
MYST1 |
NM_032188.1 |
|
|
|
|
rs4240793 |
16 |
85946457 |
0.442 |
0.192 |
-99 |
0 |
0.011 |
79791 |
FBXO31 |
NM_024735.2 |
|
|
|
|
rs333113 |
17 |
4347105 |
0.183 |
0.075 |
-99 |
0.034 |
0 |
201305 |
SPNS3 |
NM_182538.3 |
|
|
|
|
rs4968382 |
17 |
54947497 |
0.208 |
0.067 |
-99 |
0.398 |
0.389 |
79665 |
DHX40 |
NM_024612.3 |
|
|
|
|
rs1719982 |
18 |
5598808 |
0.492 |
0.167 |
-99 |
0.034 |
0.022 |
388459 |
LOC388459 |
XM_373772.3 |
|
|
|
|
rs643272 |
18 |
38442319 |
0.175 |
0.033 |
-99 |
0.273 |
0.156 |
6014 |
RIT2 |
NM_002930.1 |
|
|
|
|
rs679832 |
18 |
39208685 |
0 |
0.183 |
-99 |
0 |
0.011 |
6860 |
SYT4 |
NM_020783.2 |
|
|
|
|
rs3786467 |
18 |
53289804 |
0.025 |
0 |
-99 |
0.352 |
0.444 |
9480 |
ONECUT2 |
NM_004852.1 |
|
|
|
|
rs1823778 |
18 |
65752616 |
0.058 |
0.058 |
0 |
0.068 |
0.067 |
10666 |
CD226 |
NM_006566.1 |
|
|
|
|
rs10420077 |
19 |
35338458 |
0.167 |
0.083 |
-99 |
0.5 |
0.367 |
728091 |
LOC728091 |
XM_001126514.1 |
|
|
|
|
rs6510332 |
19 |
38222770 |
0.242 |
0.025 |
-99 |
0.455 |
0.333 |
85415 |
RHPN2 |
NM_033103.3 |
|
|
|
|
rs7253691 |
19 |
47095447 |
0.008 |
0.083 |
-99 |
0.151 |
0.267 |
9138 |
ARHGEF1 |
NM_199002.1 |
|
|
|
|
rs8110904 |
19 |
47723209 |
-99 |
-99 |
-99 |
-99 |
-99 |
634 |
CEACAM1 |
NM_001712.3 |
|
|
|
|
rs344816 |
19 |
50517466 |
0.417 |
0.05 |
0.25 |
0.093 |
0.067 |
1158 |
CKM |
NM_001824.2 |
|
|
|
|
rs2387137 |
19 |
55829472 |
0.158 |
0.042 |
-99 |
0.344 |
0.167 |
84258 |
SYT3 |
NM_032298.1 |
|
|
|
|
rs6074585 |
20 |
13223202 |
0.043 |
0.025 |
-99 |
0 |
0 |
140862 |
C20orf82 |
XM_097736.7 |
|
|
|
|
rs6141319 |
20 |
30608468 |
0.233 |
0.033 |
-99 |
0 |
0.011 |
149950 |
RP11-410N8.4 |
NM_001010976.1 |
|
|
|
|
rs6023367 |
20 |
52625318 |
0.283 |
0.083 |
-99 |
0 |
0 |
55816 |
DOK5 |
NM_018431.3 |
|
|
|
|
rs12481662 |
20 |
57451501 |
0.225 |
0.042 |
-99 |
0.102 |
0.056 |
645605 |
LOC645605 |
XM_001129785.1 |
|
|
|
|
rs6061779 |
20 |
59414462 |
0.217 |
0.1 |
-99 |
0 |
0 |
1002 |
CDH4 |
NM_001794.2 |
|
|
|
|
rs310644 |
20 |
61629948 |
0.117 |
0.075 |
-99 |
0 |
0.044 |
5753 |
PTK6 |
NM_005975.2 |
|
|
|
|
rs2823662 |
21 |
16514932 |
0.333 |
0.033 |
-99 |
0.08 |
0.044 |
388815 |
C21orf34 |
NM_001005734.1 |
|
|
|
|
rs2039248 |
21 |
17998320 |
0.258 |
0.017 |
-99 |
0.443 |
0.322 |
378826 |
C21orf114 |
NM_001012707.1 |
|
|
|
|
rs650276 |
22 |
25122236 |
0.225 |
0.017 |
-99 |
0.33 |
0.311 |
23544 |
SEZ6L |
NM_021115.3 |
|
|
|
|
rs5753625 |
22 |
30184846 |
0.342 |
0.117 |
-99 |
0.114 |
0.078 |
56478 |
EIF4ENIF1 |
NM_019843.2 |
|
|
|
|
rs1894450 |
22 |
31237721 |
0.333 |
0 |
-99 |
0.148 |
0.078 |
8224 |
SYN3 |
NM_003490.2 |
|
|
|
|
rs4824001 |
22 |
47703910 |
0.017 |
0.167 |
-99 |
0.488 |
0.456 |
25817 |
FAM19A5 |
NM_015381.3 |
|
|
|
|
rs5988072 |
X |
113797159 |
0.4111 |
0.0333 |
-99 |
0.3939 |
0.3382 |
677816 |
SNORA35 |
NR_002993.1 |
|
|
|
|
rs12125484 |
1 |
92584784 |
0.35 |
0.092 |
-99 |
0 |
0.011 |
79871 |
RPAP2 |
NM_024813.1 |
|
|
|
|
rs4768928 |
12 |
49070623 |
0.042 |
0.125 |
-99 |
0.384 |
0.467 |
121006 |
LOC121006 |
XM_926818.2 |
|
|
|
|



| File Type | application/vnd.openxmlformats-officedocument.wordprocessingml.document |
| Author | ddawson |
| File Modified | 0000-00-00 |
| File Created | 2021-02-01 |
© 2026 OMB.report | Privacy Policy